Research Areas
Ribonucleotides embedded in DNA
Ribonucleotides, the subunits of RNA, are the most abundant non-standard nucleotides found in genomic DNA. Nevertheless, there is still much to be learned about the molecular mechanisms that regulate the presence of ribonucleotides in DNA and the effects of ribonucleotides on genome stability, DNA metabolism, and human disease. In the Storici Lab, we aim to identify features and signatures of ribonucleotide incorporation, as well as molecular factors that influence the incorporation and removal of ribonucleotides from DNA, to better understand the functions and consequences of ribonucleotides on genome stability.
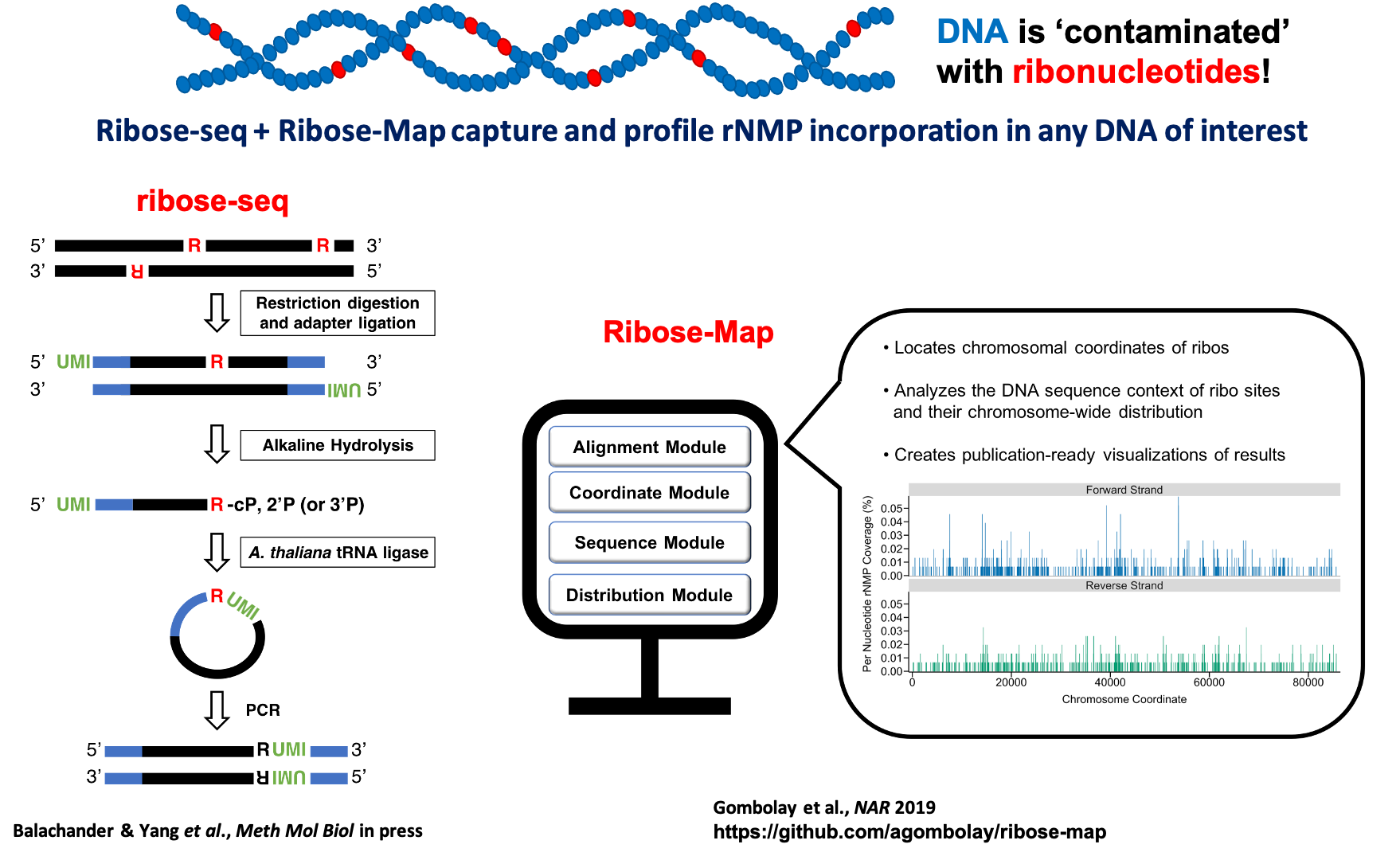
Our lab developed ribose-seq, a high-throughput sequencing technique to capture and tag ribonucleotides embedded in genomic DNA for sequencing (Koh et al., Nature Methods 2015). Ribose-seq uniquely uses alkali and AtRNL ligase from Arabidopsis thaliana to directly capture and tag the positions of embedded ribonucleotides. To analyze ribose-seq data as well as data generated from other ribonucleotide capture methods in a standardized manner, we developed Ribose-Map, a bioinformatics toolkit to map ribonucleotides embedded in genomic DNA (Gombolay et al., Nucleic Acids Research 2018). Through a series of analytical modules, researchers can use Ribose-Map to identify the exact locations of embedded ribonucleotides, study the sequence context of those ribonucleotides, and explore their genome-wide distribution. Harnessing the power of ribose-seq and Ribose-Map, we have discovered features and patterns of ribonucleotides in different yeast species (Balachander, Gombolay, Yang, Xu et al., Nature Communication 2020) and in the unicellular green alga Chlamidomonas reinhardtii (El-Sayed et al., iScience 2021). We aim to better understand the ‘language’ of ribonucleotide incorporation and its relationship to genome stability, DNA metabolism, and disease.
Relevant Publications
Gombolay et al. (2021). Mapping ribonucleotides embedded in genomic DNA to single-nucleotide resolution using Ribose-Map. Nature Protocols, in press.
Marsili et al. (2021). Gene co-expression analysis of human RNASEH2A reveals functional networks associated with DNA replication, DNA damage response, and cell cycle regulation. Biology. https://doi.org/10.3390/biology10030221
El-Sayed et al. (2021). Disproportionate presence of adenosine in mitochondrial and chloroplast DNA of Chlamydomonas reinhardtii. iScience. https://doi.org/10.1016/j.isci.2020.102005
Balachander, Gombolay, Yang, and Xu et al. (2020). Ribonucleotide incorporation in yeast genomic DNA shows preference for cytosine and guanosine preceded by deoxyadenosine. Nature Communications. https://doi.org/10.1038/s41467-020-16152-5
Gombolay, A. L., Vannberg, F. O., & Storici, F. (2019). Ribose-Map: a bioinformatics toolkit to map ribonucleotides embedded in genomic DNA. Nucleic Acids Research. doi: 10.1093/nar/gky874.
Malfatti MC, Henneke G, Balachander S, Koh KD, Newnam G, Uehara R, Crouch RJ, Storici F, Tell G (2019). Unlike the Escherichia coli counterpart, archaeal RNase HII cannot process ribose monophosphate abasic sites and oxidized ribonucleotides embedded in DNA. J Biol Chem. 30;294(35): 13061-13072. doi: 10.1074/jbc.RA119.009493
Balachander, S.*, Yang, T.*, Newnam, G., El-Sayed, W.M.M., Koh, K.D., & Storici, F. (2018) Capture of ribonucleotides in yeast genomic DNA using ribose-seq. In press in Methods in Molecular Biology ‘Yeast Systems Biology. Methods and Protocols’ (2nd ed), Springer. * Equal contribution
Malfatti, M. C.*, Balachander, S.*, Antoniali, G., Koh, K. D., Saint-Pierre, C., Gasparutto, D., Chon, H., Crouch, R. J., Storici, F.†, and Tell, G. †. Abasic and oxidized ribonucleotides embedded in DNA are processed by human APE1 and not by RNase H2. *Equal contribution; †corresponding authors. Nucleic Acids Res, 45, 11193-11212 (2017). doi: 10.1093/nar/gkx723.
Evich, M., Spring, A. M., Storici, F., and Germann, M. W. Structural impact of single ribonucleotides in DNA. Chem Bio Chem, 17(20):1968-1977 (2016).
Koh, K.D., Balachander S., Hesselberth J.R and Storici F. (2015). Ribose-seq: global mapping of ribonucleotides embedded in genomic DNA. Nature Methods, 12(3): 251-257. doi:10.1038/nmeth.3259
Shen, Y., Koh, K. D., Weiss, B. and Storici, F. (2011). Mispaired rNMPs in DNA are mutagenic and are targets of mismatch repair and RNases H. Nature Structural and Mollecular Biology, 19: 98-104. DOI: 10.1039/c4nr01794c
Shen, Y. and Storici, F. (2010). Generation of RNA/DNA hybrids in genomic DNA by transformation using RNA-containing oligonucleotides. Journal of Visualized Experiments, 45. (Video)
RNA-templated DNA break repair
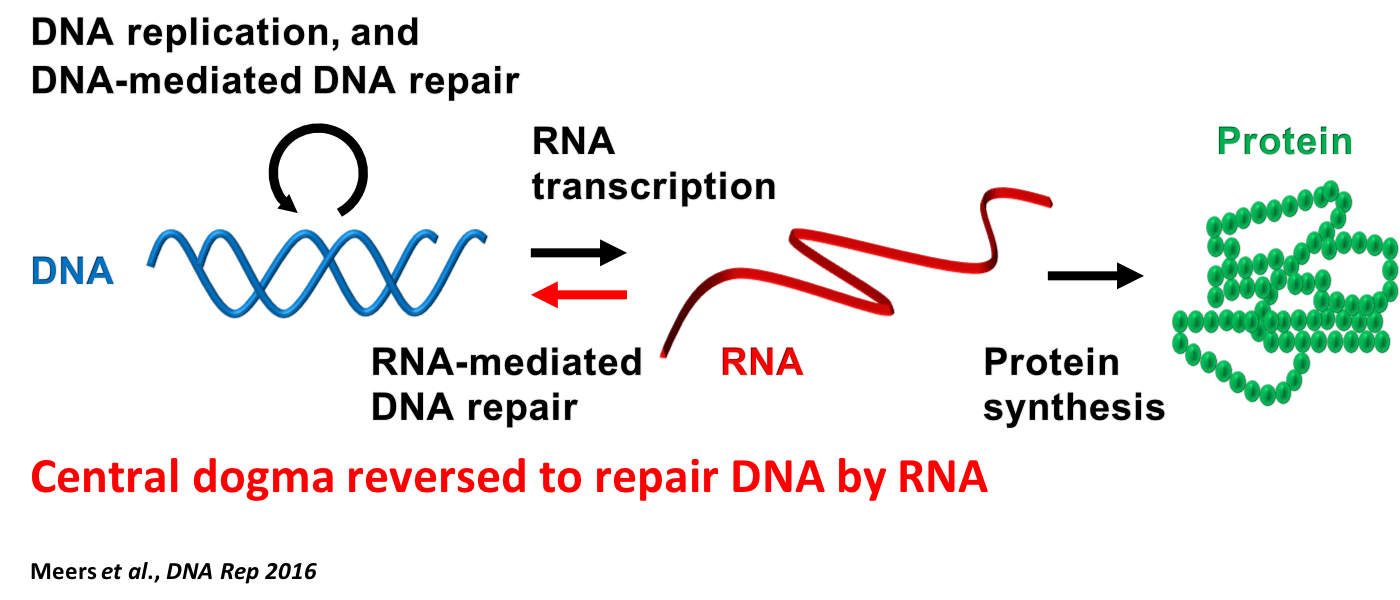
DNA is routinely damaged by various natural and environmental factors. In response, cells have evolved DNA repair mechanisms utilizing various protein factors to recognize and initiate the repair of damaged DNA. One of the most dangerous types of damage for a cell is a DNA double-stranded break (DSB), in which both DNA strands are broken along the same region. If a DSB is not precisely repaired, this can lead to mutations and chromosomal rearrangements. Using budding yeast, we found that not only homologous DNA can precisely guide the repair of a DNA DSB but also homologous RNA. We uncovered that a transcript RNA can correctly repair a DSB produced in the same DNA sequence that generated the RNA (in cis) (Keskin et al., Nature 2014; Keskin et al., RNA Biology 2016; Keskin and Storici, MicroRNA 2015). This process is repressed by endogenous ribonuclease H function, degrading RNA/DNA hybrids. In vivo and in vitro work suggests the homologous recombination enzyme Rad52 aids in repair by RNA via a mechanism of inverse strand exchange (Mazina et al., Molecular Cell 2017). Our results show that the transfer of genetic information from RNA to DNA is more general than anticipated, being not restricted to specialized RNA sequences (Meers et al., DNA Repair 2016; Michelini et al., Chemical Reviews 2018). Via a series of molecular, genetic, and biochemical assays we defined and characterized the genetic controls of three distinct mechanisms how RNA can recombine with DNA. These are: RNA-templated DSB repair (R-TDR), cDNA-templated DSB repair (c-TDR), and RNA-templated DNA modification (R-TDM), which directly modifies DNA with transcript RNA in the absence of an induced DSB and independently from Rad52 (Meers et al., Molecular Cell 2020). Now, we aim to further characterize the role of RNA in repairing DNA breaks and transferring genetic information back to DNA in different conditions and cell types, including human cells.
Relevant Publications
Marsili et al. (2021). Gene co-expression analysis of human RNASEH2A reveals functional networks associated with DNA replication, DNA damage response, and cell cycle regulation. Biology. https://doi.org/10.3390/biology10030221
Meers et al. (2020). Genetic Characterization of Three Distinct Mechanisms Supporting RNA-Driven DNA Repair and Modification Reveals Major Role of DNA Polymerase ζ. Mol Cell. https://doi.org/10.1016/j.molcel.2020.08.011
Mukherjee K, English N, Meers C, Kim H, Jonke A, Storici F, and Torres M (2020). Systematic analysis of linker histone PTM hotspots reveals phosphorylation sites that modulate homologous recombination and DSB repair. DNA Repair. https://doi.org/10.1016/j.dnarep.2019.102763
Keskin, H., Shen, Y., Huang, F., Patel, M., Yang, T., Ashley, K., Mazin, A.V., and Storici, F. (2014). Transcript-RNA-templated DNA recombination and repair. Nature 515, 436-439. doi:10.1038/nature13682
Keskin, H. and Storici, F. Defects in RNase H2 stimulate DNA break repair by RNA reverse transcribed into cDNA. MicroRNA, 4(2):109-116 (2015).
Keskin, H., Meers, C., and Storici, F. (2016). Transcript RNA supports precise repair of its own DNA gene. RNA Biology 13, 157-165.
Mazina, O.M.*, Keskin, H.*, Hanamshet, K., Storici, F., and Mazin, A.V. (2017). Rad52 Inverse Strand Exchange Drives RNA-Templated DNA Double-Strand Break Repair. Molecular Cell 67, 19-29.e13. *Equal contribution. DOI: 10.1016/j.molcel.2017.05.019
Meers, C., Keskin, H., and Storici, F. (2016). DNA repair by RNA: Templated, or not templated, that is the question. DNA Repair 44, 17-21. DOI: 10.1016/j.dnarep.2016.05.002
Michelini, F., Jalihal, A.P., Francia, S., Meers, C., Neeb, Z.T., Rossiello, F., Gioia, U., Aguado, J., Jones-Weinert, C., Luke, B., et al. (2018). From “Cellular” RNA to “Smart” RNA: Multiple Roles of RNA in Genome Stability and Beyond. Chemical Reviews 118, 4365-4403. doi: 10.1021/acs.chemrev.7b00487
DNA modification and editing
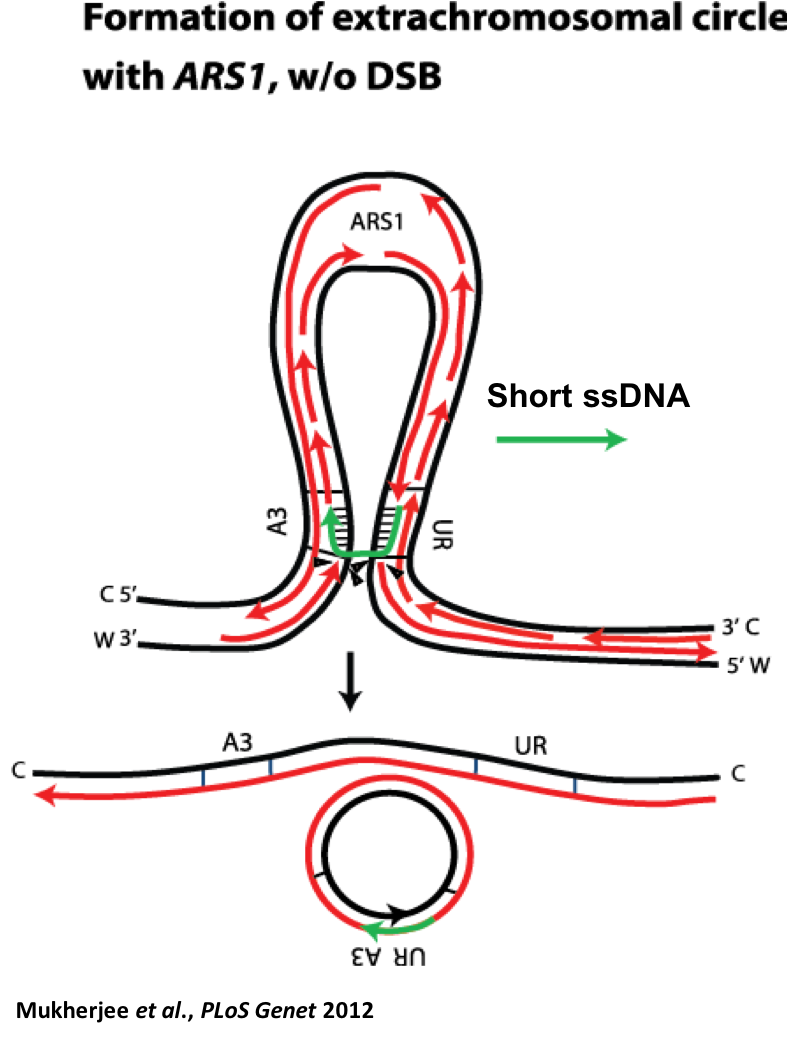
We study different mechanisms causing DNA modification using yeast and human cells. We found that small DNA fragments in the form of oligonucleotides are very recombinogenic and trigger gene amplification (small fragment-driven DNA amplification, SFDA, Mukherjee et al., PLoS Genetics 2012). SFDA events result in tandem chromosomal duplications or formation of extrachromosomal circles. These rearrangements mimic the DNA amplification structures commonly found in many cancer cells, such as the repeated units clustered at a single chromosomal locus (homogeneously staining regions, HSRs) and the circular extrachromosomal elements termed double minutes (DMs) that lack a centromere and telomeres but retains the ability to replicate autonomously.
We aim to improve the efficiency and safety of gene editing using yeast and human cell systems. We have optimized conditions and protocols for the delitto perfetto approach for in vivo site-specific mutagenesis with a DSB and distant from a DSB using oligonucleotides in budding yeast (Stuckey and Storici, In: Laboratory Methods in Enzymology 2013; Katz and Storici, Methods in Molecular Biology 2014). We developed the ‘gene collage’ system for easy integration of multiple recombinant/heterologous DNA segments into yeast chromosomal DNA (Stukey et al., Methods in Molecular Biology 2011). Because DNA DSB systems used to stimulate gene correction efficiency often pose a threat due to their high likelihood to generate off-target DNA damage, nicking enzymes represent attractive tools for safer genome engineering. We have provided new mechanistic insights into the process of nick-induced DNA recombination and on the function of nicking enzymes in gene targeting (Katz et al., PLoS One 2014).
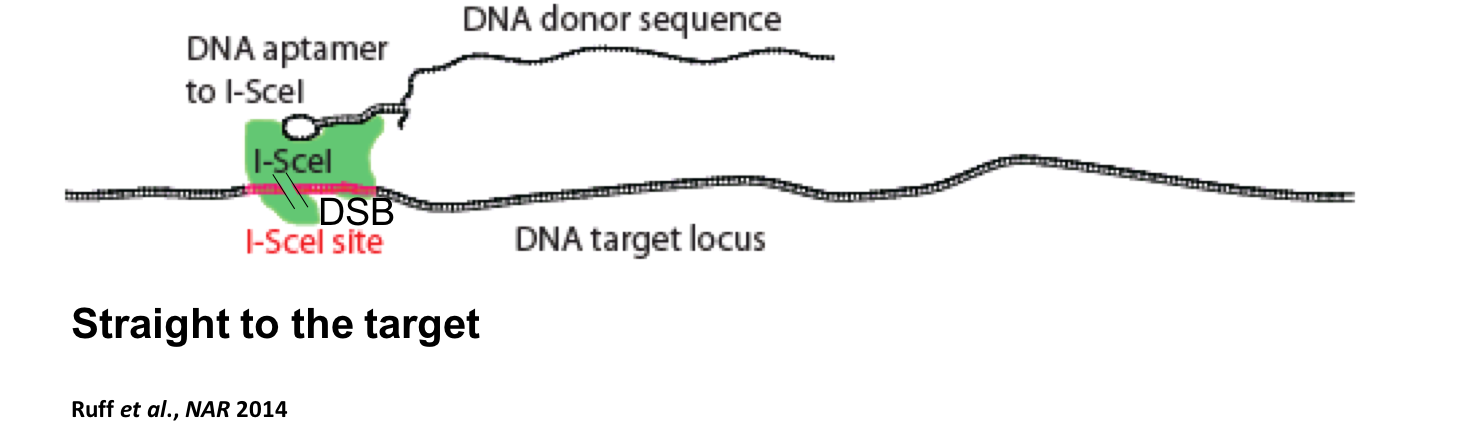
Furthermore, we focus our research on the donor DNA necessary to make the desired genetic modification during the process of gene editing. We developed a novel gene targeting approach, aptamer-guided gene targeting (AGT), in which we bound the homing endonuclease I-SceI by a DNA aptamer fused to the donor DNA of choice, to target the donor DNA to a desired genetic locus located next to an I-SceI cut site (Ruff et al., Nucleic Acids Research 2014). We found that the AGT approach increases the efficiency of gene targeting by guiding an exogenous donor DNA into the vicinity of the site targeted for genetic modification. This study shows that DNA aptamers can be exploited to increase donor DNA availability, and thus promote the transfer of genetic information from a donor DNA molecule to a desired genetic locus (Ruff and Storici In: Genome Editing 2016).
Relevant Publications
Ruff, P. and Storici, F. Genome editing by aptamer-guided gene targeting (AGT). In: “Genome Editing: The Next Step in Gene Therapy” Edited by T. Cathomen, M. Hirsch, and M. Porteus. American Society of Gene and Cell Therapy and Springer publishing. Vol 895, pp 111-124 (2016).
Ruff, P., Koh, K. D., Keskin, H, Pai, R. and Storici, F. Aptamer-guided gene targeting in yeast and human cells. Nucleic Acids Research 42, No. 7 e61 doi: 10.1093/nar/gku101 (2014).
Storici, F., Editor. (2014) Gene Correction: Methods and Protocols – Methods in Molecular Biology; Humana Press, New York, NY; Vol. 1114.
Katz, S. S., Gimble, F. S. and Storici, F. To nick or not to nick: comparison of I-SceI single- and double-strand break-induced recombination in yeast and human cells. PLoS One. 9 (2), e88840 (2014).